Dr. Ana Eulalio
RNA: the missing link in bacterial pathogen-host interactions

Institute for Molecular Infection Biology
University Wurzburg
Josef-Schneider-Str. 2/D15
D-97080 Wurzburg
Germany
Email: ana.eulalio@uni-wuerzburg.de
Homepage
Research
Our main goal is to unravel hidden roles of RNA molecules in the context of bacterial pathogen-host interactions, by focusing on the impact of bacterial infection on host cell RNA metabolism, as well as the reciprocal effect of these pathways on the life cycle of pathogenic bacterial, issues that remain largely unexplored. Our work will be focused on three main lines of research:
1. Interplay between host cell microRNAs and bacterial infection
Despite previous studies, by us and others, showing that bacterial pathogens (e.g. Salmonella enterica, Helicobacter pylori) induce significant changes in the mammalian host microRNA expression profile, the generation of a comprehensive atlas of microRNAs regulated as a consequence of bacterial infection is clearly missing. Therefore, a major focus of our research is the identification of host microRNAs regulated upon infection with representative bacterial pathogens, as well as the characterization of the mechanisms by which bacteria modulate host cell microRNAs. Our ultimate aim is to determine if a microRNA signature exists for infection with different bacteria, and to establish whether microRNAs are regulated in a cell-type specific manner. We are also working on the identification of host cell microRNAs that regulate bacterial infection.
2. Impact of bacterial pathogens on host cell RNA metabolism
The relationship between bacterial infections and RNA granules, in particular P-bodies (also known as mRNA processing bodies) and stress-granules, is another aspect of the bacterial-host interaction for which very little information is available. Considering that the formation and stability of RNA granules is strictly dependent on the cellular RNA metabolism, any perturbation induced by bacterial pathogens on RNA granules is most likely a consequence of their effect on host cell RNA metabolism. We aim to determine whether P-bodies, stress-granules, and/or their protein components are implicated in the regulation of RNA metabolism during bacterial infections, which will be important to determine whether and how bacteria interfere with RNA-related processes in their hosts. The reciprocal effect of RNA metabolism on bacterial infection will also be evaluated. Overall, with these studies we aim to characterize the impact of bacteria on cellular mRNA processing, stability and surveillance pathways.
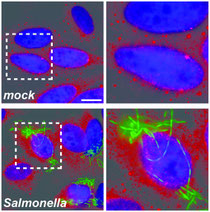
Figure Legend
Salmonella infection induces P-body disassembly. HeLa cells were mock-treated or infected with Salmonella enterica serovar Typhimurium expressing GFP. P-bodies were detected by staining the cells with anti-GW182 antibody (red channel). The regions highlighted by the white squares are enlarged on the rightmost panels. Scale bar, 10 µm.
3. Effect of bacterial non-coding RNAs on key host intracellular pathways
One of the most intriguing questions that we aim to address is whether bacteria have developed a system(s) to actively introduce RNAs inside host cells to modulate key cellular functions. This hypothesis is particularly pertinent taking into consideration that bacteria have evolved very efficient secretion systems for proteins and that a number of non-coding RNAs with, so far, unknown function have been identified in bacteria.
Group members
Clivia Lisowski - PhD student
Dr. Claire Maudet-Crépin - Postdoc
Dorothee Sennhenn - Technician
Malvika Sharan - PhD student
Ushasree Sunkavalli - PhD student
Publications within BioSysNet
Maudet C, Mano M, Sunkavalli U, Sharan M, Giacca M, Förstner KU, Eulalio A (2014). Functional high-throughput screening identifies the miR-15 microRNA family as cellular restriction factors for Salmonella infection. Nat Commun. [Epub ahead of print]
Maudet C, Mano M, Eulalio A (2014). MicroRNAs in the interaction between host and bacterial pathogens. FEBS Lett. [Epub ahead of print]
Eulalio A, Schulte L, Vogel J (2012). The mammalian microRNA response to bacterial infections. RNA Biol. 9(6):742-750.
Publications before BioSysNet
Schulte L, Eulálio A, Mollenkopf H, Reinhardt R, Vogel J (2011)
Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family
EMBO J 30(10):1977-1989
Eulálio A, Fröhlich K, Mano M, Giacca M, Vogel J (2011)
A candidate approach implicates the secreted Salmonella effector protein SpvB in P-body disassembly
PLoS One 6(3):e17296
Heale BS, Eulálio A, Schulte L, Vogel J, O'Connell MA (2010)
Analysis of A to I editing of miRNA in macrophages exposed to Salmonella
RNA Biol 7:116-122
Eulálio A, Tritschler F, Izaurralde E (2009)
The GW182 protein family in animal cells: new insights into domains required for miRNA-mediated gene silencing
RNA 15:1433-1442
Eulálio A, Helms S, Fritzsch C, Fauser M, Izaurralde E (2009)
A C-terminal silencing domain in GW182 is essential for miRNA function
RNA 15:1067-1077
Eulálio A, Tritschler F, Büttner R, Weichenrieder O, Izaurralde E, Truffault V (2009)
The RRM domain in GW182 proteins contributes to miRNA-mediated gene silencing
Nucleic Acids Res 37(9):2974-2983
Tritschler F, Braun J, Eulálio A, Truffault V, Coles M, Izaurralde E, Weichenrieder O (2009)
Structural basis for the mutually exclusive anchoring of P-body components EDC3 and Tral to the DEAD Box Protein DDX6/Me31B
Mol Cell 33:661-668
Eulálio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E (2009)
Deadenylation is a widespread effect of miRNA regulation
RNA 15:21-32
Tritschler F, Eulálio A, Helms S, Schmidt S, Coles M, Weichenrieder O, Izaurralde E, Truffault V (2008)
A similar mode of interaction enables Trailer Hitch and EDC3 to associate with DCP1 and Me31B in distinct protein complexes
Mol Cell Biol 28:6695-6708
Jinek M, Eulálio A, Lingel A, Helms S, Conti E, Izaurralde E (2008)
The C-terminal region of Ge-1 presents conserved structural features required for P-body localization
RNA 14:1991-1998
Eulálio A, Huntzinger E, Izaurralde E (2008)
Argonaute and GW182 interaction is essential both for miRNA-mediated translational repression and mRNA decay
Nat Struct Mol Biol 15:346-353
Nunes-Correia I, Rodriguez JM, Eulálio A, Carvalho AL, Citovsky V, Simões S, Faro C, Salas ML, Pedroso de Lima MC (2008)
African swine fever virus p10 protein exhibits nuclear import capacity and accumulates in the nucleus during viral infection
Vet Microbiol 130:47-59
Eulálio A, Huntzinger E, Izaurralde E (2008)
Getting to the Root of miRNA-Mediated Gene Silencing
Cell 132:9–14
Tritschler F, Eulálio A, Truffault V, Hartmann MD, Helms S, Schmidt S, Coles M, Izaurralde E, Weichenrieder O (2007)
A divergent Sm-fold in EDC3 proteins mediates DCP1-binding and P-body targeting
Mol Cell Biol 27:8600-8611
Eulálio A, Rehwinkel J, Stricker M, Huntzinger E, Yang SF, Doerks T, Dorner S, Bork P, Boutros M, Izaurralde E (2007)
Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing
Genes Dev 21:2558-2570
Dorner S, Eulálio A, Huntzinger E, Izaurralde E (2007)
Delving into the diversity of silencing pathways. Symposium on MicroRNAs and siRNAs: Biological Functions and Mechanisms
EMBO Rep 8:723 - 729
Eulálio A, Nunes-Correia I, Salas J, Salas ML, Simões S, Pedroso de Lima MC (2007)
African swine fever virus p37 structural protein is localized in nuclear foci containing the viral DNA at early post-infection times
Virus Res 130:18-27
Eulálio A, Behm-Ansmant I, Schweizer D, Izaurralde E (2007)
P-body formation is a consequence, not the cause of RNA-mediated gene silencing
Mol Cell Biol 27:3970-3981.
Eulálio A, Behm-Ansmant I, Izaurralde E (2007)
P-bodies: at the crossroads of post-transcriptional pathways
Nat Rev Mol Cell Biol 8:9–22
Eulálio A, Nunes-Correia I, Carvalho AL, Faro C, Citovsky V, Salas J, Salas ML, Simões S, Pedroso de Lima MC (2006)
Nuclear export of African swine fever virus p37 protein occurs through two distinct pathways and is mediated by three independent signals
J Virol 80:1393-1404
Eulálio A, Nunes-Correia I, Carvalho AL, Faro C, Citovsky V, Simões S, Pedroso de Lima MC (2004)
Two African Swine Fever virus proteins derived from a common precursor exhibit different nucleo-cytoplasmic transport activities
J Virol 78:9731-9739
Nunes-Correia I, Eulálio A, Nir S, Pedroso de Lima MC (2004)
Caveolae as an additional route for Influenza Virus endocytosis in MDCK cells
Cell Mol Biol Lett 9:47-60
Nunes-Correia I, Eulálio A, Nir S, Düzgünes N, Ramalho-Santos J, Pedroso de Lima MC (2002)
Fluorescent probes for monitoring virus fusion kinetics: Comparative evaluation of reliability
Biochim Biophys Acta 1561:65-75



